Organisation - Unimore |
|
|
|
|
 UNIMORE - University of Modena and Reggio Emilia (www.unimore.it) is an Italian public University with a longstanding tradition, founded in 1175. Located in one of Europe's wealthiest and most dynamic regions, the Region Emilia-Romagna, UNIMORE is based in the two nearby cities of Modena and Reggio Emilia. The university ranked among the top 8 universities in Italy for its level of research, according to the most recent assessment of the Italian Ministerial Committee for the Evaluation of Research (CIVR). It is composed of 14 Departments, offering a wide range of degree programs at the undergraduate level, right up to doctoral studies in most disciplinary areas, from the humanities and social sciences to engineering and technology, and from physical and natural sciences to medicine and life sciences. UNIMORE, which has just over 19,000 students including 3,500 postgraduates, is large enough to offer all the facilities one would expect from a major university but small enough to retain a personal and friendly learning environment. It offers state-of-the-art facilities with internationally sponsored research activities and with the two Technopoles of Modena and Reggio Emilia; UNIMORE is in fact part of the High Technology Regional Network (HTN). UNIMORE - University of Modena and Reggio Emilia (www.unimore.it) is an Italian public University with a longstanding tradition, founded in 1175. Located in one of Europe's wealthiest and most dynamic regions, the Region Emilia-Romagna, UNIMORE is based in the two nearby cities of Modena and Reggio Emilia. The university ranked among the top 8 universities in Italy for its level of research, according to the most recent assessment of the Italian Ministerial Committee for the Evaluation of Research (CIVR). It is composed of 14 Departments, offering a wide range of degree programs at the undergraduate level, right up to doctoral studies in most disciplinary areas, from the humanities and social sciences to engineering and technology, and from physical and natural sciences to medicine and life sciences. UNIMORE, which has just over 19,000 students including 3,500 postgraduates, is large enough to offer all the facilities one would expect from a major university but small enough to retain a personal and friendly learning environment. It offers state-of-the-art facilities with internationally sponsored research activities and with the two Technopoles of Modena and Reggio Emilia; UNIMORE is in fact part of the High Technology Regional Network (HTN). The Crop Production Group (www.cropsci.unimore.it) based at the Department of Life Sciences of UNIMORE (www.dsv.unimore.it) was established in 2005 by Nicola Pecchioni. The Group is now composed of a group leader (prof. N. Pecchioni), one researcher (dr. E. Francia), one technician (dr. J.A. Milc), two postdocs, two PhD students and one foreign fellow. It primarily conducts research on both crop and model plants to improve their productivity and end-use quality. Since 2008 it has given a substantial contribution to the Interdepartmental Centre for Genomic Research (www.cgr.unimore.it) of UNIMORE. Other than NGS facilities of CGR, the group has an experimental field in Gattatico (Reggio Emilia) for phenotyping and yield trials.
Genomic research of the group is focused on the biological mechanisms limiting crop production under biotic and abiotic stress conditions. Barley (Hordeum vulgare) is main target species. The Crop Production Group (www.cropsci.unimore.it) based at the Department of Life Sciences of UNIMORE (www.dsv.unimore.it) was established in 2005 by Nicola Pecchioni. The Group is now composed of a group leader (prof. N. Pecchioni), one researcher (dr. E. Francia), one technician (dr. J.A. Milc), two postdocs, two PhD students and one foreign fellow. It primarily conducts research on both crop and model plants to improve their productivity and end-use quality. Since 2008 it has given a substantial contribution to the Interdepartmental Centre for Genomic Research (www.cgr.unimore.it) of UNIMORE. Other than NGS facilities of CGR, the group has an experimental field in Gattatico (Reggio Emilia) for phenotyping and yield trials.
Genomic research of the group is focused on the biological mechanisms limiting crop production under biotic and abiotic stress conditions. Barley (Hordeum vulgare) is main target species.
 Barley Genetics and Genomics
Barley Genetics and GenomicsFrom the beginning of its scientific activity, the group studied barley under abiotic stress (cold and drought) condition as well as the genomics of barley adaptation to Mediterranean environments. The development of a Nure x Tremois (NT) population, conceived as idea in late 90's for QTL mapping has been unexpectedly fruitful. We firstly described the Fr-H1+Fr-H2 2-loci system of resistance to frost in barley (2004), and we firstly in barley indicated the CBF genes as candidates of Fr-H2 (2004 and 2007). In collaboration with CRA-GPG we have developed a barley genetic map with more than 500 markers (2014). We independently developed other Nure-derived genetic materials such as large segregating populations and QTL-NILs. Other (few) QTLs for frost tolerance have been added through a GWAS approach (2013).
As regards the study of barley yield adaptation to the environment, In collaboration with the barley scientific communit we contributed with the NT population to the discovery of barley "Vrn" vernalization genes (2005), and to the discovery of barley powdery mildew resistance meta-QTLs (2010). It was demonstrated and quantified by us how vernalization+flowering time genes are 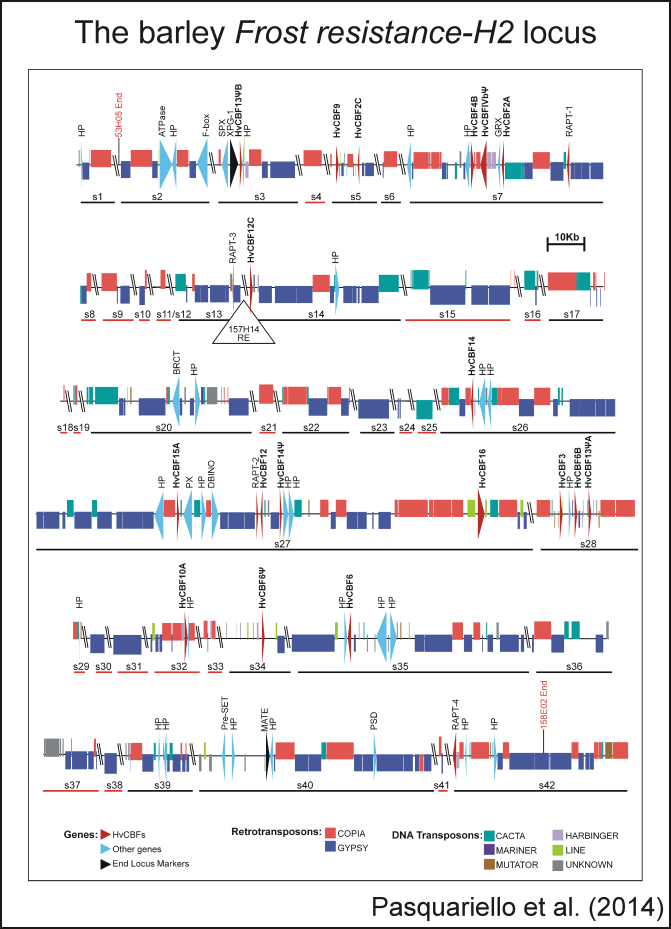 The current research interests are the elucidation of the genetic and molecular bases of grain yield and tolerance to low temperature and drought. The construction of an integrated physical-genetic map of a region of barley chromosome 5H that harbour a major QTL of frost resistance (Fr-H2) and a CBF-gene cluster is one of the main recent achievements. Massive parallel de novo sequencing allowed structural and functional characterization of the locus. The research is complemented by expression studies of the CRT/DRE (CBF) binding factors gene family, residing at Fr-H2, as well as of their putative regulators. The current research interests are the elucidation of the genetic and molecular bases of grain yield and tolerance to low temperature and drought. The construction of an integrated physical-genetic map of a region of barley chromosome 5H that harbour a major QTL of frost resistance (Fr-H2) and a CBF-gene cluster is one of the main recent achievements. Massive parallel de novo sequencing allowed structural and functional characterization of the locus. The research is complemented by expression studies of the CRT/DRE (CBF) binding factors gene family, residing at Fr-H2, as well as of their putative regulators.Other Research Interests relevant for Barley and Triticeae
The Crop Production group constituted in 2007 a first Italian platform for genetics and genomics of the model species Brachypodium distachyon. A Brachypodium genetic map has been constructed at UNIMORE and three QTLs of tolerance to false brome rust (Puccinia brachypodii) identified. Brachypodium genomic response to the fungal pathogen has been described by transcript profiling, and functional genomics results compared to genetic ones for the identification of resistance causal genes.
The group contributed to the realization of a physical map of wheat chromosome 5A, under the frame of a national effort. As a part of the Regional High Technology Network (HTN), the Crop Production group is active in the Net-Lab BIOGEST- SITEIA, that deals with research transfer to the agro-food industry, in which the group is leading the section devoted to technology transfer to Seed Companies. One of the main activities is the development, implementation and improvement of a comprehensive database integrating the molecular and phenotypic information for wheat, barley, and rice, to help regional and national cereal breeders in the MAS (marker-assisted-selection) process (www.cerealab.org ). Recent selected publications
Pasquariello M, Barabaschi D, Himmelbach A, Steuernagel B, Ariyadasa R, Stein N, Gandolfi F, Tenedini E, Bernardis I, Tagliafico E, Pecchioni N, Francia E (2014). The barley Frost resistance-H2 locus, Functional & Integrative Genomics, OnlinePub:10.1007/s10142-014-0360-9
Tondelli A., E. Francia, A. Visioni, J. Comadran, A.M. Mastrangelo, T. Akar, A. Al-Yassin, S. Ceccarelli, S. Grando, A. Benbelkacem, F.A. van Eeuwijk, W.T.B. Thomas, A.M. Stanca, I. Romagosa, N. Pecchioni (2014). QTLs for barley yield adaptation to Mediterranean environments in the 'Nure' x 'Tremois' biparental population. Euphytica 197(1): 73-86. Visioni A, Tondelli A, Francia E, Pswarayi A, Malosetti M, Russell J, Thomas W, Waugh R, Pecchioni N, Romagosa I, Comadran J (2013). Genome-wide association mapping of frost tolerance in barley (Hordeum vulgare L.), BMC Genomics 14(1): 424 Galiba G., E.J. Stockinger, E. Francia, J. Milc, G. Kocsy and N. Pecchioni (2013). Freezing tolerance in the Triticeae. In: "Translational Genomics for Crop Breeding" (Eds Varshney RK and Tuberosa R), Volume II: Abiotic Stress, Quality and Yield. John Wiley & Sons, USA pp 99-124. Comadran J, Kilian B, Russell J, Ramsay L, Stein N, Ganal M, Shaw P, Bayer M, Thomas W, Marshall D, Hedley P, Tondelli A, Pecchioni N, Francia E, Korzun V, Walther A, Waugh R (2012). Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley, Nature Genetics, 44: 1388-1392 Barbieri M, TC. Marcel, R.E. Niks, E. Francia, M. Pasquariello, V. Mazzamurro, D.F.. Garvin, and N. Pecchioni (2012). QTLs for resistance to the leaf rust Puccinia brachypodii in the model grass Brachypodium distachyon L. Genome 55: 152-163 Pecchioni N., J. Milc, M. Pasquariello and Enrico Francia (2012). Barley: Omics Approaches for Abiotic Stress Tolerance. In: "Improving Crop Resistance to Abiotic Stress", N Tuteja, Sarvajeet S. Gill, Antonio F. Tubercio and Renu Tuteja (Editors), Wiley-Blackwell, Wiley-VCH Verlag GmbH & Co., Germany. Vol. 2 pp. 779-884 Francia E, Tondelli A, Rizza F, Badeck FW, Li Destri Nicosia O, Akar T, Grando S, Al-Yassin A, Benbelkacem A, Thomas WTB, van Eeuwijk F, Romagosa I, Stanca AM, Pecchioni N (2011). Determinants of barley grain yield in a wide range of Mediterranean environments, Field Crops Research, 120: 169-178 Milc J., Sala A, Bergamaschi S and Pecchioni N. (2011) A genotypic and phenotypic information source for marker-assisted selection of cereals: the CEREALAB Database. Database, Vol. 2011, Article ID baq038, doi:10.1093/database/baq038 |
|
Barley Genome Net - Organisation - Unimore |
|
|
|
|
|
|
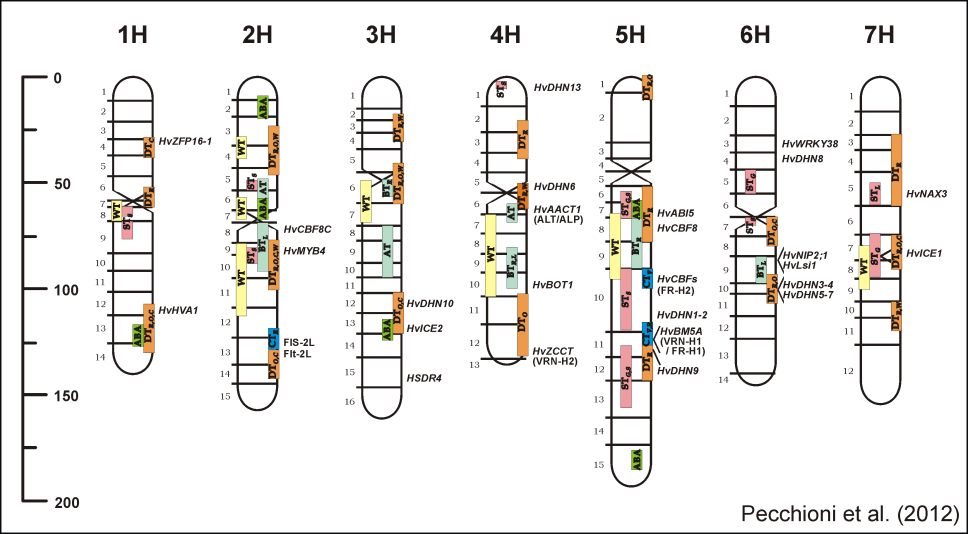 determinants of barley grain yield in Mediterranean environments (2011 and 2013). In 2012, the group also contributed to the cloning of the EPS2 QTL responsible of earliness per se, as one of major drivers of yield adaptation of spring and winter barleys in Europe.
determinants of barley grain yield in Mediterranean environments (2011 and 2013). In 2012, the group also contributed to the cloning of the EPS2 QTL responsible of earliness per se, as one of major drivers of yield adaptation of spring and winter barleys in Europe.


