Organisation - KVL |
|
|
|
|
|
The Royal Veterinary and Agricultural University (KVL) was founded in 1856. KVL is the only Danish university specialising in the agricultural and veterinarian area and the only place where veterinarians, agronomists, horticulturists and MSc's in forestry and agricultural economists are educated. KVL is also one the few universities in Denmark where MSc's in food science and technology are educated. Research, both basic and applied, is carried out at KVL within natural science, veterinary medicine, domestic animals, environment, agriculture, plant culture, landscape architecture, organic farming, forestry, food and human nutrition. The central campus area covers more than 36 acres and includes, among others, a veterinary clinic, greenhouses, research laboratories, lecture rooms, offices and the Danish Veterinary and Agricultural Library. Besides this, KVL also has an arboretum and three experimental farms within 20 km from the central campus. 3,500 students, of whom 120 are foreigners and 400 are PhD students, attend KVL who employs 1,600 people and has a budget of DKK 960 million including DKK 300 million from external funding. During 2005 the plant biotechnology group at Risø National Laboratory was merged with KVL to form the Molecular Plant Breeding group in the Plant and Soil Laboratory. The Molecular Plant Breeding group is localized within the Department of Agricultural Sciences at KVL. This significantly strengthen KVLs capacity in providing courses in molecular plant breeding and research aiming at Plant for the Future. The group now counts over 25 scientist, post docs, phd-students and master thesis students. It covers five research areas within major crop plants (barley, wheat and rice) Plant Breeding (Sven Bode Andersen, Professor), Defence Mechanism (Hans Thordal-Christensen, Research leader), Respiration (I. Max Møller, Research Professor), Diversity, Drought and Resistance (Ahmed Jahoor, senior research scientist) and Plant Quality (Søren K. Rasmussen, Research leader).
Research relevant to BarleyGenomeNet (selection)Breeding for disease resistanceIn order to strengthen disease resistance in crops, the Department of Agricultural Sciences co-operates with Danish and Swedish plant breeders. The Department develops advanced breeding lines with new and efficient resistance genes and develops molecular markers to select for the resistance in the progeny of the crosses made with the resistance donor lines. This includes linkage mapping of the respective genes and in the case of quantitative resistance also the QTL mapping strategies. The molecular breeding group has especially a long and outstanding experience with barley leaf diseases, such as powdery mildew, leaf rust and scald and disposes of excellent genetic resources from wild barley, Hordeum spontaneum with resistance genes that have never been used in breeding. In the frame of this project, sources of resistance to powdery mildew and leaf rust the wild barley Hordeum spontaneum from the "Fertile Crescent" has been employed. For scald resistance, landraces from East Asian origin are used. Crosses between the donor lines and European cultivars have been developed in close co-operation with Danish as well Swedish plant breeders in order to map the genes for resistance and developed markers for marker assisted selection.
A TILLING population of barleyThe Tilling population is EMS-mutated; the wild type is the Danish variety 'Lux'. From ca. 10.000 M3 lines, DNA has been isolated in close co-operation with ICARDA, the International Center for Agricultural Research in the Dry Areas, one of the 15 CGIAR centers and located in Aleppo, Syria. We have pooled DNA from 10 mutant lines and perform PCR reactions on the pools with primer pairs designed for our genes of interest. The primers of one pair have different dyes attached. After the PCR reaction we add a further step of denaturation and annealing. This steps results in the formation of small loops, if mutations are present in one of the lines in the pool. Hereafter, we add CEL I to the reaction, an endonuclease cleaving at those sequence mismatches. A fragment analysis on a semi-automated sequencer shows either only the full-length product, if no mutation was present in the pooled lines or additional bands corresponding to the product from the primer to site of the mutation(s). While this technique originally was designed to clarify the function of known sequences, an additional benefit is the selection of mutants with a desired trait. Therefore, we select candidate genes we expect to have consequences for the trait we are interest in, analyse them by TILLING and test the phenotype with appropriate procedures. The results are lines with changed trait expression that are not classified as genetic modified plants. At present, efforts are devoted to identify mutants for drought tolerance as well as for quality.
Drought tolerance (A. Jahoor)In co-operation with ICARDA, the Department for Agricultural Sciences uses molecular breeding tools to enhance the drought tolerance of barley. While the development of DNA-markers for drought-tolerance related traits through QTL mapping has a long history in this co-operation, the use of a mutant population through TILLING and the screening of natural variation for specific genes through EcoTILLING, both recently developed techniques, is a fairly new approach in our research. For QTL mapping, three way crosses have been developed involving Syrian landraces, wild barley Hordeum spontaneum and European cultivars in order to combine the drought tolerance from landraces, disease resistance from wild barley and yield potential from European cultivars. All field experiments are conducted at ICARDA, whereas mapping is carried out at Royal Veterinary and the Agricultural University, Copenhagen. Phytate metabolism (S. Rasmussen, KVL)We study the biosynthesis of phytate and its deposition during seed development as the main source of stored seed phosphorus. Mutants have been selected to enable elucidation of the biosynthetic pathway and key enzymes and genes are studied. The storage of phytate with iron and zinc and other minerals is of particular interest. We are also studying the brake down of phytate by endogenous phytase and phosphatases into inositol releasing phosphorus during germination. For the society phytate influence many aspects which relates to phosphorus being a limiting natural resource and phosphorus as a pollution from domestic animal production. Phytate may potentially be anticarcinogenic but also hinders the bioavailability of the iron and zinc from food and feed. Besides mutational breeding, we use TILLING and transgenic plants in pursuing our goals.
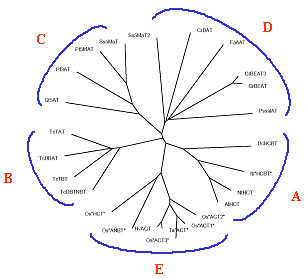
Hordatine: agmatine coumaroyltransferase (ACT) is a key enzyme in antifungal cinnamic acid amide biosynthesis (S. Rasmussen, KVL)Hydroxycinnamoylagmatines are direct precursor of hordatines, which are antifungal compounds found to be highly abundant in the young barley seedling (3). The hordatines seem to be confined to the genus Hordeum as preformed infection inhibitors, and recent studies indicate that the synthesis of hydroxycinnamoylagmatine derivatives are induced in response to fungal infection of the leaves. Additionally, hydroxycinnamoylagmatine derivatives have been found in wheat, and histochemical staining of epidermal leaf tissue confirms that these compounds might accumulate in cereals in general as a response to fungal infection. The function(s) of hydroxycinnamoylagmatine derivatives in plants is not known but may include cell wall fortification, restriction of pathogeningress, and cytotoxicity to the invading pathogen. The purified transferases are all cytosolic proteins and large family of structurally related acyltransferases produce a diverse numbers of natural products including aroma, flavour and scent.
|
|
Barley Genome Net - Organisation - KVL |
|
|
|
|
|
|